
Yogesh Verma
I'm Yogesh, a Doctoral Researcher at QQML group at Aalto University with Prof. Vikas Garg, Prof. Samuel Kaski, and Dr. Markus Heinonen focusing on physics-inspired deep learning for modeling dynamical systems, (geometric and topological) deep learning, generative models and robust bayesian inference.
I am also a recipient of Nokia Scholarship, Encouragement Grant by FFTP, and a DAAD AI-Net Fellow.
Email | LinkedIn | GitHub | Google Scholar
Selected publications
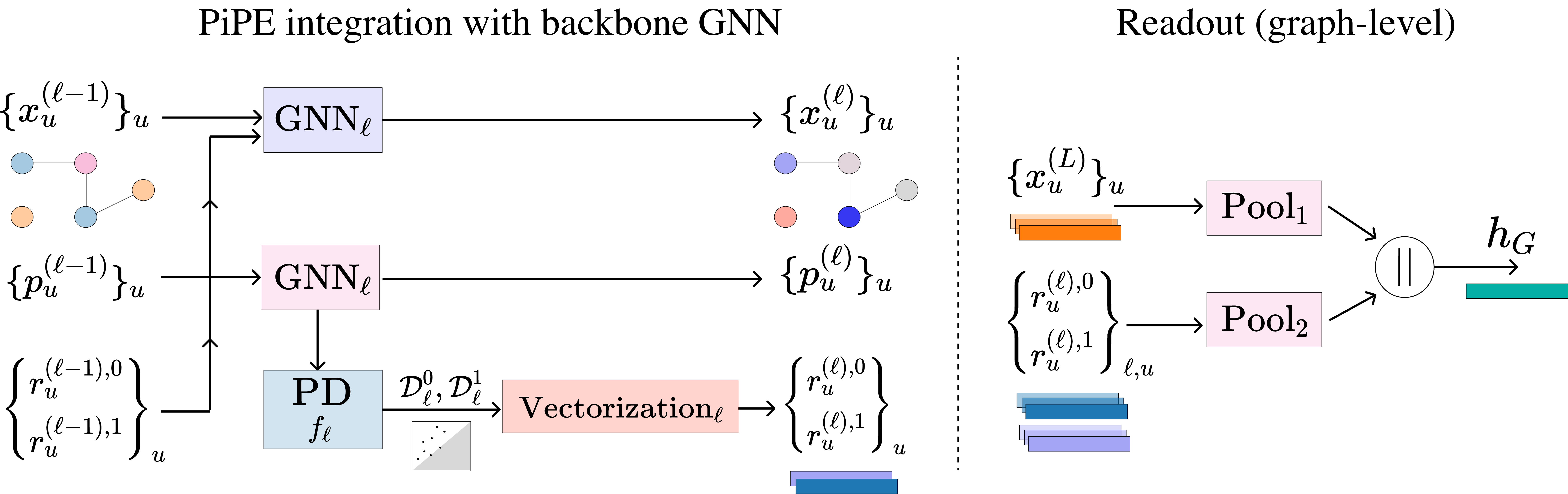
Positional Encoding meets Persistent Homology on Graphs
ICML, 2025
Message-passing GNNs struggle to capture global structures like connectivity and cycles. We theoretically compare two enhancements—Positional Encoding (PE) and Persistent Homology (PH)—showing that each has unique strengths and limitations. Building on this, we propose PiPE, a novel method combining both, which is provably more expressive and achieves strong results across diverse graph learning tasks.
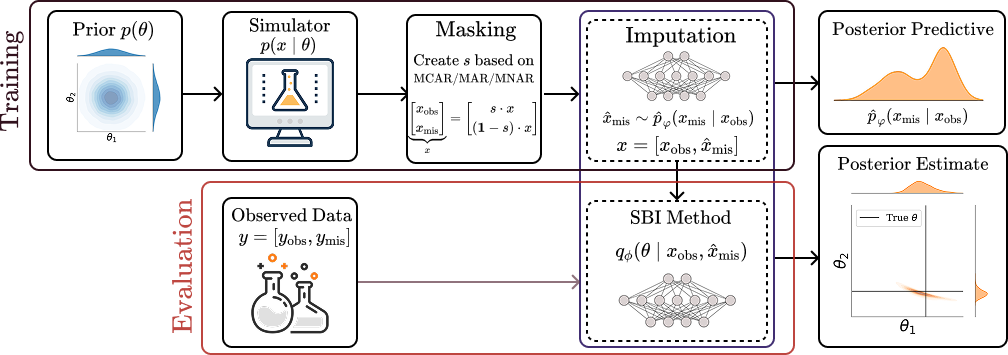
Robust Simulation-Based Inference under Missing Data via Neural Processes
ICLR, 2025
We investigates the robustness of NPE based methods under missing data observations in simulation-based inference (SBI). We are the first to identify the inherent bias introduced by missing observations in these methods. To address this, we propose a novel method that enhances the robustness of NPE methods under missing data, achieving state-of-the-art performance and providing more reliable inference.
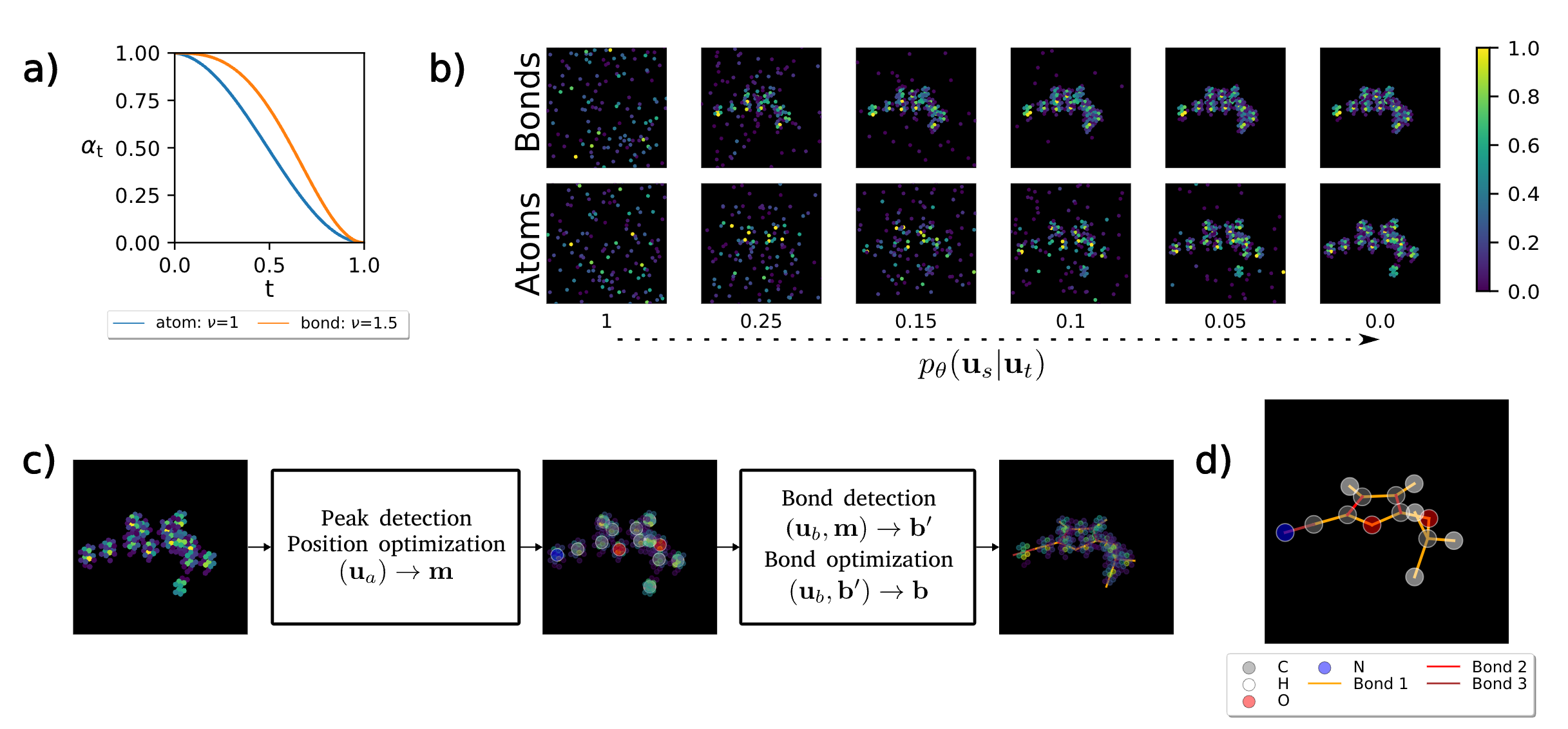
E(3)-equivariant models cannot learn chirality: Field-based molecular generation
ICLR, 2025
We show that existing molecule generative models inherently disregard chirality—a critical geometric property affecting drug safety and efficacy. To overcome this, we propose a novel field-based representation with reference rotations that relax strict rotational symmetry. Our method captures all molecular geometries, including chirality, while matching the performance of E(3)-equivariant models on standard benchmarks.
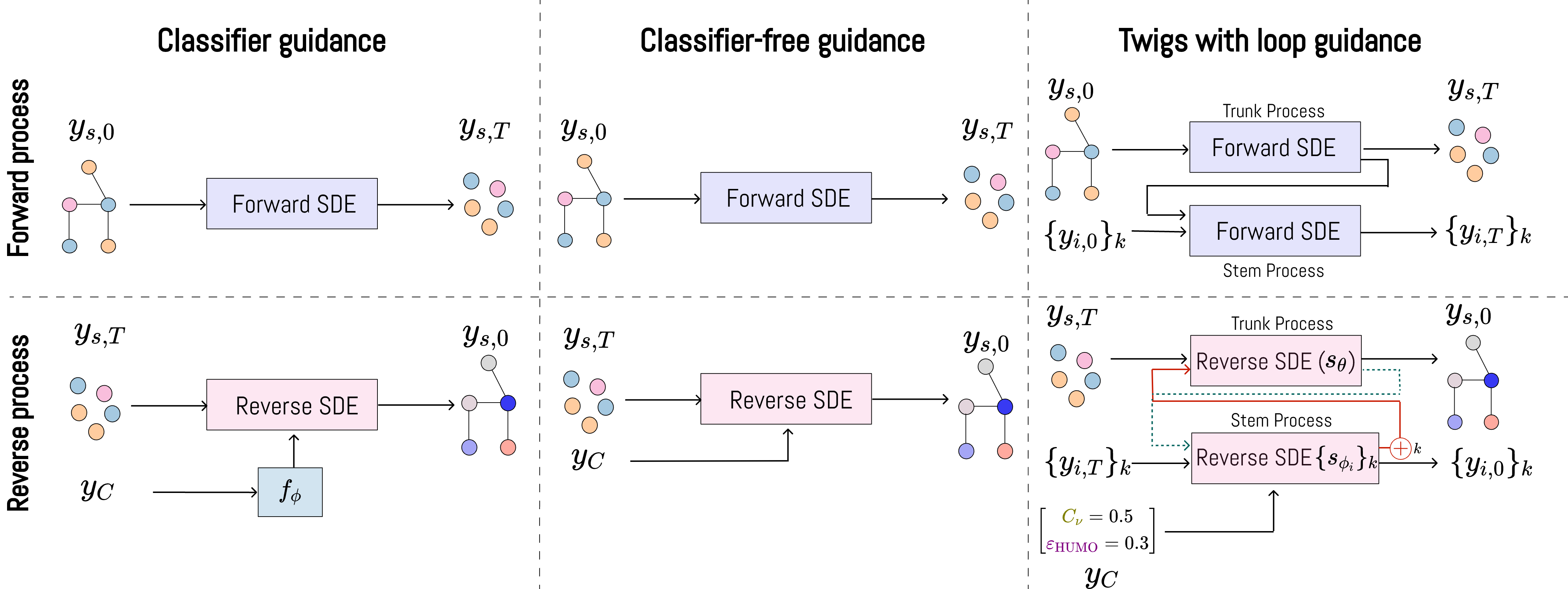
Diffusion Twigs with Loop Guidance for Conditional Graph Generation
NeurIPS, 2024
We proposes a novel hierarchical generative framework for conditional generation of drug molecules with state-of-the-art performance. Using the proposed method, we can design better drug molecules with finer control over the properties of the molecule.
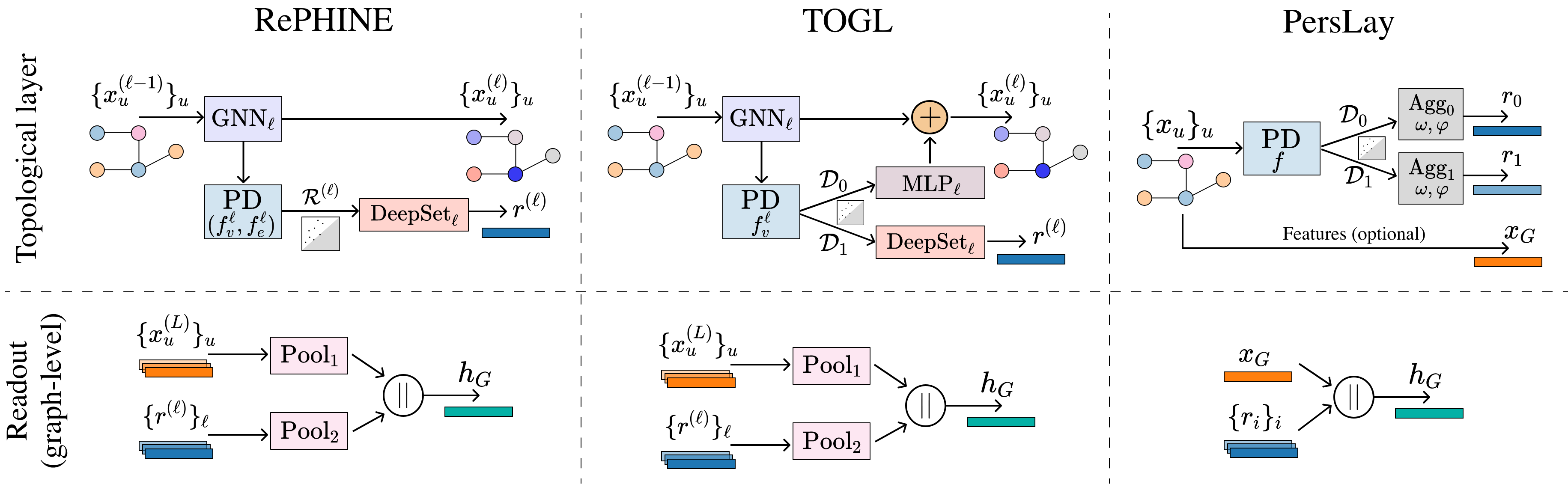
Topological Neural Networks go Persistent, Equivariant and Continuous
ICML, 2024
We introduce TopNets as a broad framework that subsumes and unifies various methods in the intersection of GNNs/TNNs and PH such as (generalizations of) RePHINE and TOGL. TopNets can also be readily adapted to handle geometric complexes. Theoretically, we show that PH descriptors can provably enhance the expressivity of simplicial message-passing networks. Empirically, TopNets achieve strong performance across diverse tasks.
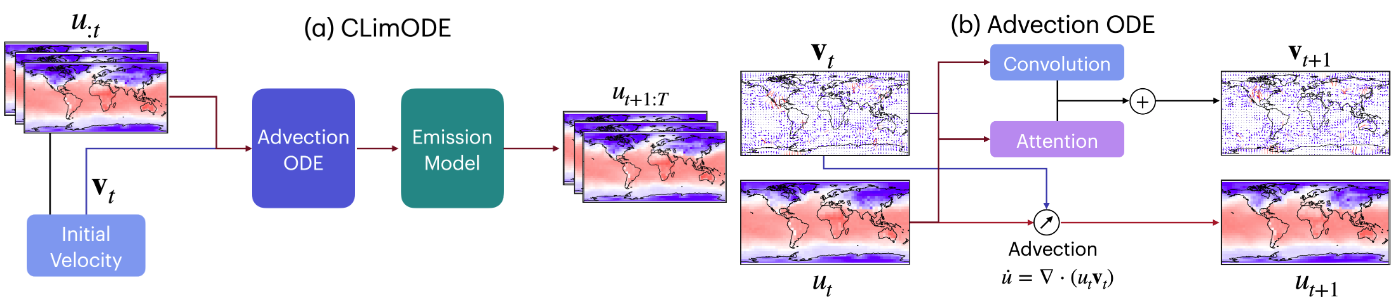
ClimODE: Climate and Weather Forecasting With Physics-informed Neural ODEs
ICLR, Oral, 2024
We propose ClimODE, a spatiotemporal continuous-time process that implements a key principle of advection from statistical mechanics. ClimODE models precise weather evolution with value-conserving dynamics, learning global weather transport as a neural flow, which also enables estimating the uncertainty in predictions. Our approach outperforms existing data-driven methods in global and regional forecasting with an order of magnitude smaller parameterization.
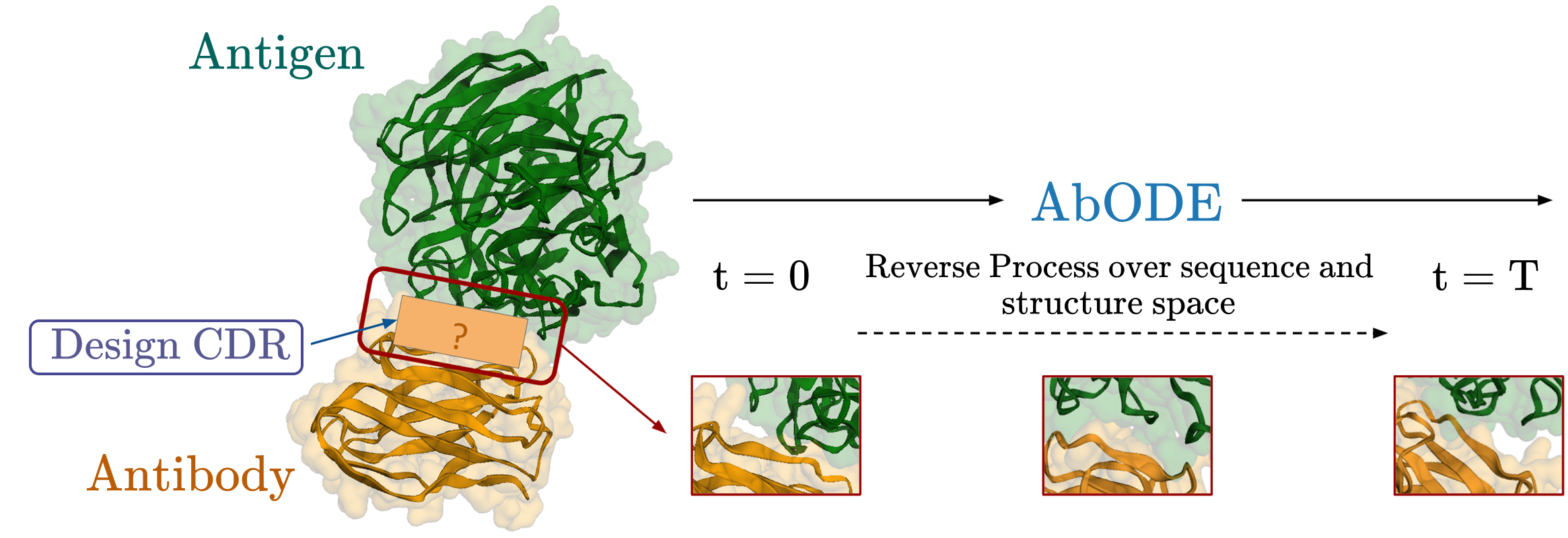
AbODE: Ab initio Antibody Design using Conjoined ODEs.
ICML, 2023
We propose a new generative model AbODE that extends graph PDEs to accommodate both contextual information and external interactions. AbODE uses a single round of full-shot decoding and elicits continuous differential attention that encapsulates and evolves with latent interactions within the antibody as well as those involving the antigen. The proposed model significantly outperforms existing methods on standard metrics across benchmarks.
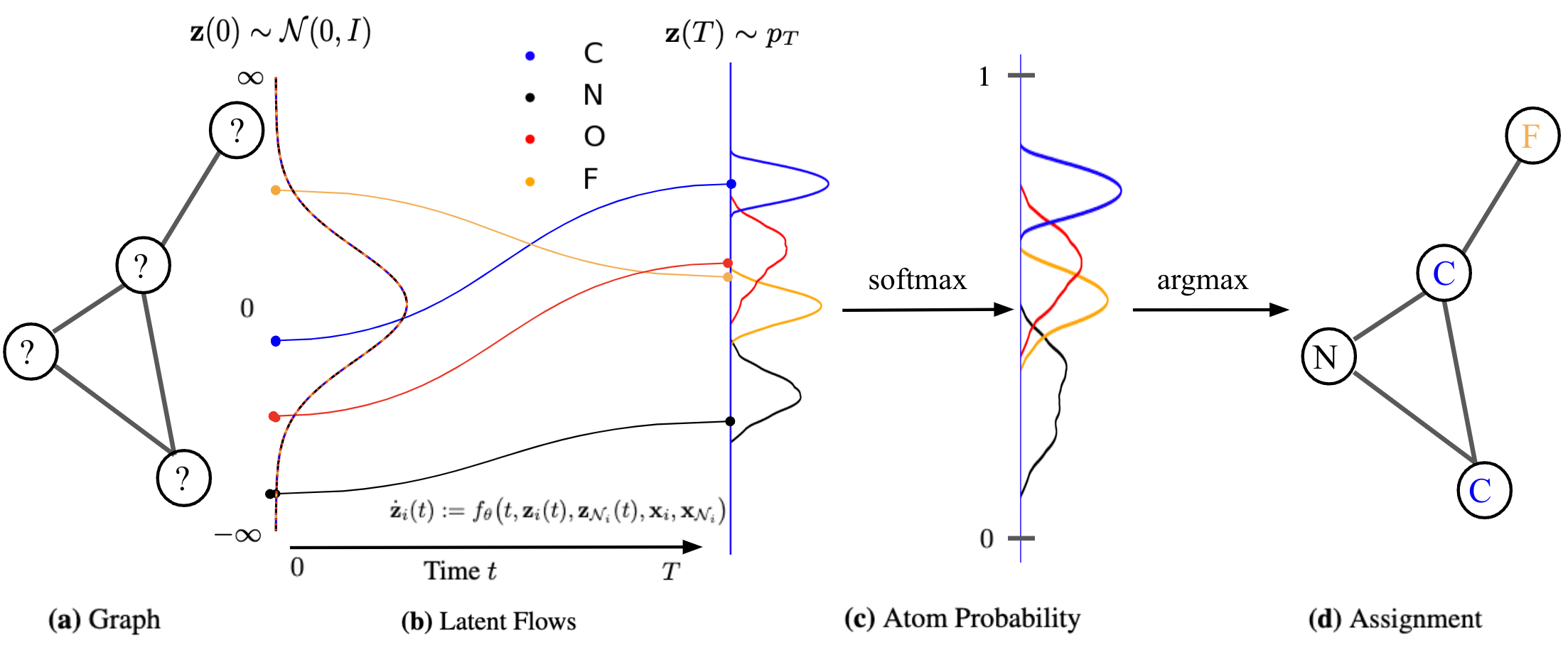
Modular Flows: Differential Molecular Generation
NeurIPS, 2022
We propose a continuous normalizing E(3)-equivariant flows, based on a system of node ODEs, that repeatedly reconcile locally toward globally aligned densities. Our models can be cast as message-passing temporal networks, and result in superlative performance on the tasks of density estimation and molecular generation.
Invited Talks
- Math+ML+X Seminar, Tsinghua University, 2025
- ELLIS Advanced Probabilistic ML Seminar, ELLIS Helsinki, 2024
- Workshop on Physics-Informed Learning, ELISE Wrap Up Conference, ELISE, ELLIS, 2024
- AI4Science Talks, University of Stuttgart and NEC Labs Europe, 2024
- Research Seminar, MLLS Institute, Denmark, 2023
Reviewing
- NeurIPS, ICML, ICLR, AISTATS, JMLR, AABI